---
title: Illumina
date: 2026-01-01
published-title: Created
date-modified: last-modified
title-block-banner: "#212529"
# toc: true
# toc-location: left
toc-title: "Contents"
execute:
eval: false
format:
html:
code-tools:
source: true
toggle: true
---
## Sequencing basics
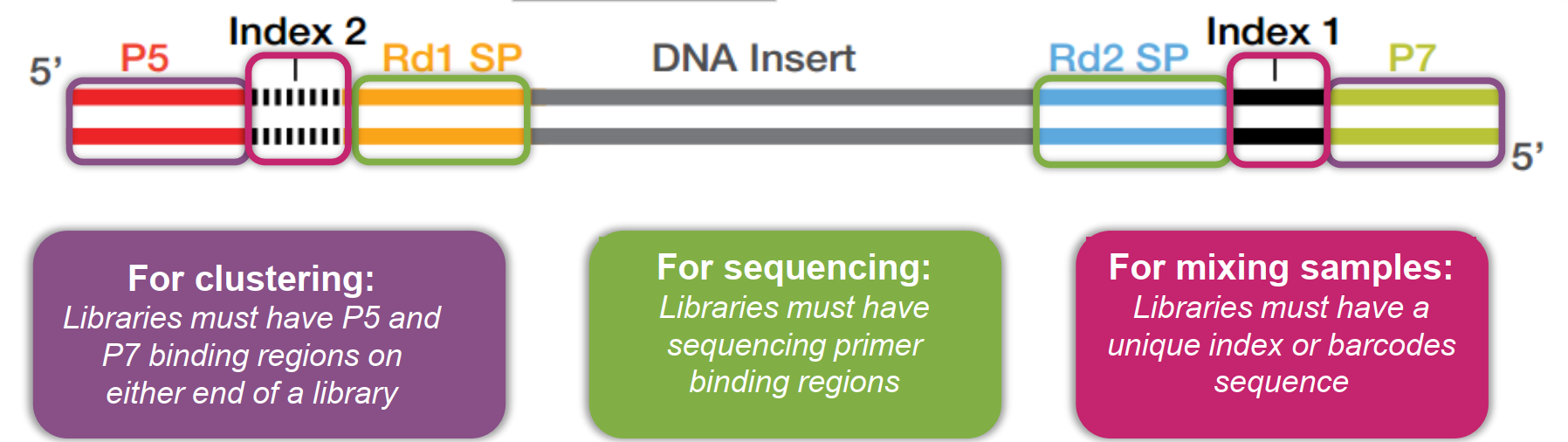{width=600px}
- Clusters: Groups of DNA strands positioned closely together. Each clustter represents thousands of copies of the same DNA fragment in a 1-2 micron spot
- Flowcell: A thick glass slide with channels or lanes. Cluster generation and sequencing occur here. Each lane is randomly coated with a lawn of oligos that are complementary to library adapters.
- Random Flow cell
- Patterned Flow cell
- Reads: The sequences of nucleotides (A, T, C, G) generated from the DNA fragments during sequencing.
- Lanes: Individual channels on a flowcell that can be used for separate samples or experiments.
- Indexing: Adding unique sequences (barcodes) to DNA fragments to identify different samples in a single sequencing run.
- Adapters: Short DNA sequences attached to the ends of DNA fragments to facilitate binding to the flowcell and initiation of sequencing.
- Paired-end sequencing: Sequencing both ends of a DNA fragment to provide more information and improve accuracy.
- Coverage: The average number of times a nucleotide is read during sequencing, indicating the depth of sequencing.
- Read length: The number of nucleotides in a single read generated by the sequencer.
- Throughput: The total amount of data generated by a sequencing run, often measured in gigabases (Gb) or terabases (Tb).
- Multiplexing: Combining multiple samples in a single sequencing run using unique indexes to save time and cost.
- Demultiplexing: The process of separating mixed sequencing data back into individual samples based on their unique indexes.
- Quality scores: Numerical values assigned to each nucleotide in a read, indicating the confidence in the accuracy of that base call.
- Phred score: A specific type of quality score that represents the probability of an incorrect base call, commonly used in sequencing data analysis.
- FASTQ format: A text-based file format that stores both nucleotide sequences and their corresponding quality scores.
- BCL files: Binary files generated by Illumina sequencers that contain raw base call data and quality scores before conversion to FASTQ format.
## Quality scores
- Illumina uses Phred quality scores (Q scores) to represent the accuracy of each base call in sequencing data.
- The Q score is calculated using the formula: Q = -10 log10(P), where P is the probability of an incorrect base call.
- Higher Q scores indicate higher confidence in the accuracy of the base call.
- For example:
- Q10: 90% accuracy (1 in 10 chance of error)
- Q20: 99% accuracy (1 in 100 chance of error)
- Q30: 99.9% accuracy (1 in 1000 chance of error)
- Q40: 99.99% accuracy (1 in 10,000 chance of error)
- Q50: 99.999% accuracy (1 in 100,000 chance of error)
## Illumina 5-base
1. Map: Align reads to the reference.
2. Call: Decide if a "T" is a 5th base (5mC) or a mutation.
3. Overlay: Compare those sites to a database of known genes and CpG islands.
4. Compare: Look for differences between samples to find "Biological Hits."
